Strain-Level Phage-Bacteria Interactions Shape Honeybee Gut Microbial Dynamics
November 5, 2025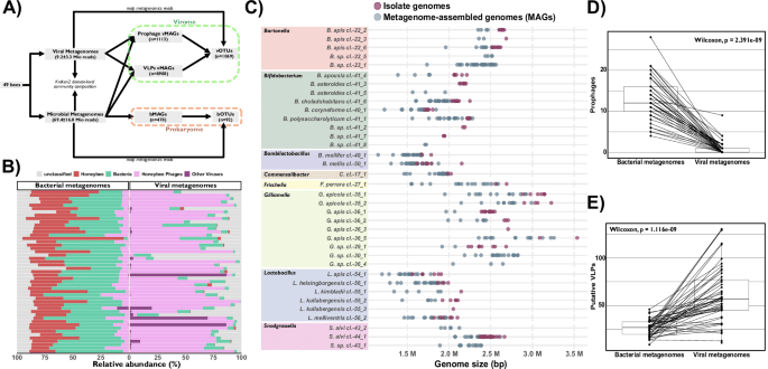
The PBIN network is highly modular, consisting of nine modules and a nested structure within those modules, with modularity aligning to core bee gut genera and limited cross-genus host ranges among phages.
Network structure is nested within modules for most modules, a pattern confirmed by isolate-only PBIN analyses; modularity is driven by bacterial phylogeny and phage co-clustering within modules.
Mantel tests reveal a significant correlation between bacterial and viral compositions, strongest at the strain level and when taxa share the same module, indicating ecologically meaningful PBIN units.
New findings show that strain-level interactions drive honeybee gut microbial dynamics, and the PBIN’s modular-nested structure explains how phage and bacterial diversity relate across individual bees.
There is a strong positive link between within-module bacterial diversity (across phylogeny and nucleotide variation) and within-module viral diversity (vOTU richness), reinforcing PBIN-driven diversity coupling.
The study analyzed 49 individual Apis mellifera bees using paired shotgun metagenomics to capture both viral and bacterial fractions and to construct a PBIN across strains and modules.
Bacterial communities show low beta-diversity at genus/species levels across bees but high strain-level variability, while viral communities vary markedly at the vOTU level between individuals.
Overall, phages are recognized as key shapers of bacterial communities, with evidence and remaining gaps particularly at the bacterial strain level within proposed host interaction networks.
The dataset includes 478 reconstructed bMAGs forming 53 bOTUs across 49 bees and 10,021 vMAGs clustered into 1,069 vOTUs, with most vOTUs predicted to infect core gut bacteria.
A majority of vMAGs are linked to at least one bacterial genome, with hundreds of vOTUs predicted to infect core gut bacteria, inferred through CRISPR spacers and genome homology.
Summary based on 1 source
