Revolutionizing Drug Discovery: Embracing Protein Dynamics and Flexibility for Better Predictions
November 6, 2025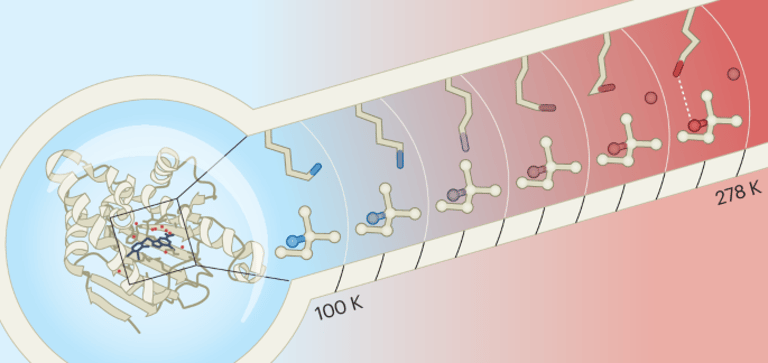
The story centers on methodological advances in capturing protein dynamics, including room-temperature X-ray crystallography, multi-temperature and multi-conformer modeling (qFit, FLEXR, mmCIF ensembles, qFit 3), and ensemble refinement as tools to reveal native-like flexibility.
Historical work showed that temperature, cryocooling, and crystal packing can distort protein structures, underscoring the need to reinterpret static images in drug design.
The overarching takeaway is a shift toward embracing conformational diversity as a native feature, not a flaw, to improve structure-based drug discovery.
At the core, the collection emphasizes improving drug discovery by accounting for protein flexibility and conformational ensembles in structural biology.
Practically, studies discuss artifacts from cryocooling, the importance of sampling heterogeneity, and methods that better predict catalysis and binding mechanisms in enzymes and drug targets.
Temperature effects, hydration networks, interfacial water, and dynamic binding sites are highlighted as factors that influence ligand binding predictions and docking accuracy.
Technological progress—from serial crystallography to neutron/X-ray methods and synchrotron-enabled approaches—broadens access to dynamic structural information.
Milestones include Fraser et al. on room-temperature ensembles, Keedy et al. on hidden backbone conformations, Burnley et al. on ensemble refinement, and 2022–2024 work on high-resolution dynamics and mmCIF encoding.
Summary based on 1 source