Revolutionary CondenSeq Method Unveils Secrets of Nuclear Protein Condensates
June 21, 2025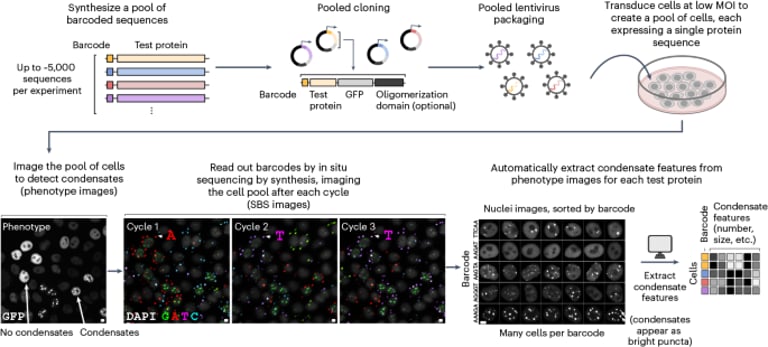
A novel high-throughput method called CondenSeq has been developed to analyze and decode protein sequences that contribute to the formation of nuclear condensates.
This method employs a pooled imaging technique to evaluate around 14,000 synthetic protein sequences for their ability to form condensates.
The study identifies multiple classes of nuclear condensates, each driven by distinct sequence determinants.
To support their research, the authors utilized several publicly accessible databases for sequence design and analysis, including MobiDB, LLPSDB, Disprot, and Phasepro.
Images from the study have been uploaded to the Bioimage Archive, accessible via the provided DOI link, under accession number S-BIAD1738.
Predictions for the large library sequences generated by CondenSeq are available on Zenodo, offering additional resources for researchers.
Processed data for all protein sequences is included in the Supplementary Data files, which can be accessed through links in the article's supplementary materials.
Additional image analysis code and examples are available on GitHub, aiding researchers in their own analysis efforts.
The GitHub repository also includes code modifications of previously published work to enhance analysis capabilities.
The references list provides key literature on biomolecular condensates and phase separation, offering a comprehensive background for the study.
Overall, this research enhances the understanding of biomolecular condensates, which are critical to cellular biochemistry.
Results from the application of CondenSeq provide insights into the relative impact of various sequence features across different contexts.
Summary based on 2 sources
Get a daily email with more Science stories
Sources
Nature • Jun 20, 2025
High-throughput method for decoding the protein sequence determinants of condensates