Breakthrough in DNA-Targeting: New Computational Method Designs Highly Specific DNA-Binding Proteins
September 12, 2025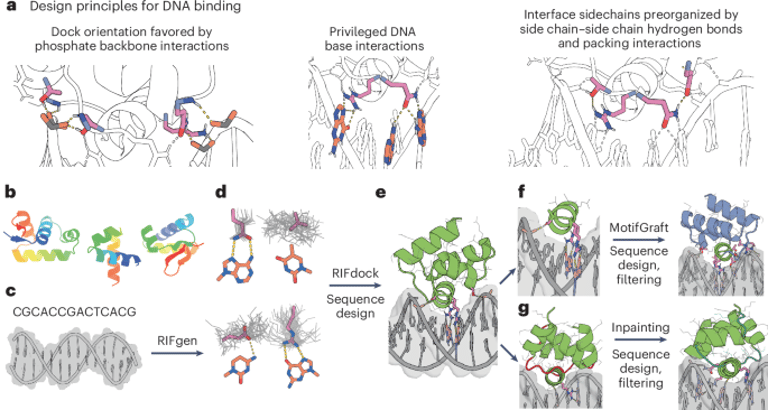
The design process involved sampling diverse scaffold structures from metagenome data, docking these against DNA targets to optimize interactions, and using deep-learning tools like ProteinMPNN and Rosetta for sequence and structural refinement, including side-chain preorganization.
Some of the designed DBPs showed high specificity for their target sequences, confirmed through competition assays and mutational footprinting, demonstrating the accuracy of the design process.
Over 300,000 potential DBP designs were generated across different target sets, with yeast surface display and fluorescence-activated cell sorting (FACS) used to identify candidates that bind their intended DNA sequences.
Researchers have developed a computational approach to design sequence-specific DNA-binding proteins (DBPs) using small helical domains, mainly helix–turn–helix (HTH) scaffolds, to target specific DNA sequences.
Experimental testing revealed that 44 of these candidate designs bound specifically to their target DNA sequences, with affinities from 30 to 500 nanomolar, and mutations at key interface residues disrupted binding, confirming the models.
Crystallographic analysis of one DBP in complex with its DNA target showed close agreement with the design model, with key hydrogen bonds and water-mediated interactions aligning with predictions, supporting the structural validity.
This integrated approach combining de novo docking, deep learning, and structural validation can produce highly specific, novel DNA-binding proteins, offering new tools for gene regulation, editing, and diagnostics beyond existing systems like zinc fingers, TALEs, and CRISPR-Cas.
Compared to native protein–DNA complexes, many of the designed DBPs employ novel interfaces and docking orientations, expanding the known docking space and recognizing sequences not typically targeted by natural DBPs.
Overall, this work demonstrates a successful pipeline for designing compact, sequence-specific DNA binders with high precision and specificity, validated through structural and biochemical characterization.
Summary based on 1 source
Get a daily email with more Science stories
Source

Nature • Sep 12, 2025
Computational design of sequence-specific DNA-binding proteins