ProKAS: Revolutionary Biosensor Maps Kinase Activity in Living Cells, Advancing Drug Discovery
November 14, 2025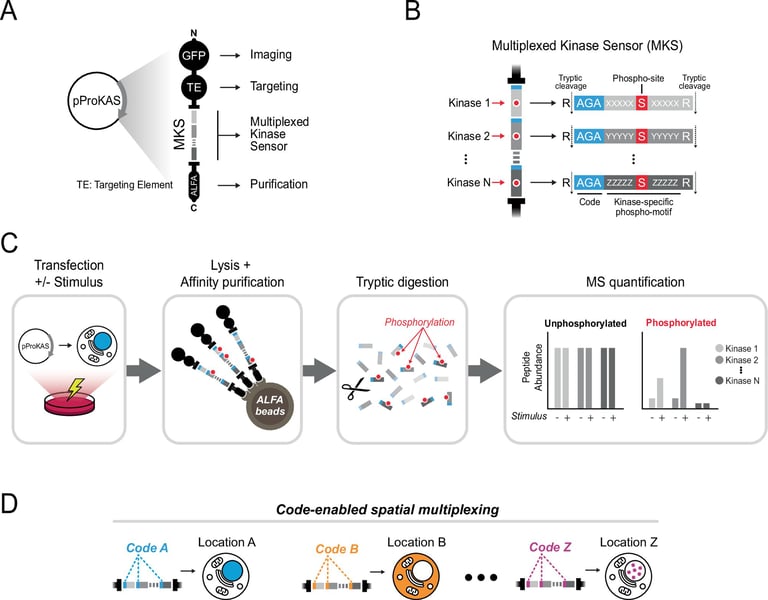
ProKAS introduces a mass-spectrometry–based, multiplexed biosensor system that maps kinase activity inside living cells with spatial resolution.
The system uses barcoded substrate peptides to report where and when kinases act, enabling simultaneous monitoring of multiple kinases across cellular regions, including nuclear areas.
Cornell researchers developed ProKAS as a novel platform that preserves unmodified sensor abundance while tracking phosphorylation changes, enabling sub-stoichiometric sensing without depleting sensors.
A proof-of-principle set focuses on DNA damage response kinases ATR, ATM, CHK1, with expansion to include a pan-CDK sensor and a PPM1D phosphatase sensor for broader network monitoring.
The authors design RS peptides from endogenous substrates and validate ATR sensors using FANCD2 S717 with CPT/HU treatments, confirming specificity via ATR inhibitors.
Sensors are linked to localization tags to achieve spatial data, enabling nucleus-versus-cytosol analysis of phosphorylation without sensor loss.
The platform can be adapted to study other human kinases and may accelerate drug action studies through rapid, high-throughput kinase activity screening.
Spatial data show constitutive phosphorylation is higher in the nucleus than in the cytosol, aligning with nuclear DNA damage signaling while enabling compartment-specific activity assessment.
A multiplexed DDR ProKAS sensor set monitors ATR, ATM, and CHK1 responses to CPT and HU, and gauges inhibitor potency and selectivity, including ATM-specific AZD0156.
Future directions include integrating ProKAS with computational design, expanding peptide libraries, and clarifying how kinases shape cell behavior and cancer-related responses.
The sensor focus on DDR kinases ATR, ATM, and CHK1 reveals region-specific activity differences not detectable before.
Localization to nuclei or cytosol is achieved via targeting elements, enabling precise compartmental analysis of kinase activity.
Each sensor peptide yields distinctive tryptic phosphopeptides for MS detection, allowing simultaneous measurement of multiple kinase activities within a single ProKAS construct.
The core sensor system (MKS) comprises a tandem array of 10–15 amino acid substrates, framed by barcodes and a localization element to direct subcellular targeting.
A de novo design approach uses PSPA data and a genetic algorithm to identify CHK1 sensors with high specificity, supported by CHK1 inhibition validation.
ProKAS enables high-throughput analysis, demonstrated with 36 samples in a 30-minute mass spectrometry run, with plans to scale to hundreds or thousands of samples.
Kinetic analyses reveal distinct activation timelines for ATM, ATR, and CHK1 under CPT and HU conditions, underlining ProKAS’s quantitative capabilities.
Overall, ProKAS offers a versatile, high-throughput platform for mapping kinase signaling networks with multiplexed, spatiotemporal resolution, with broad implications for drug discovery and kinase biology.
Summary based on 2 sources