Asgard Archaea: Key Ancestors Shaping Eukaryotic Complexity Before LECA
January 14, 2026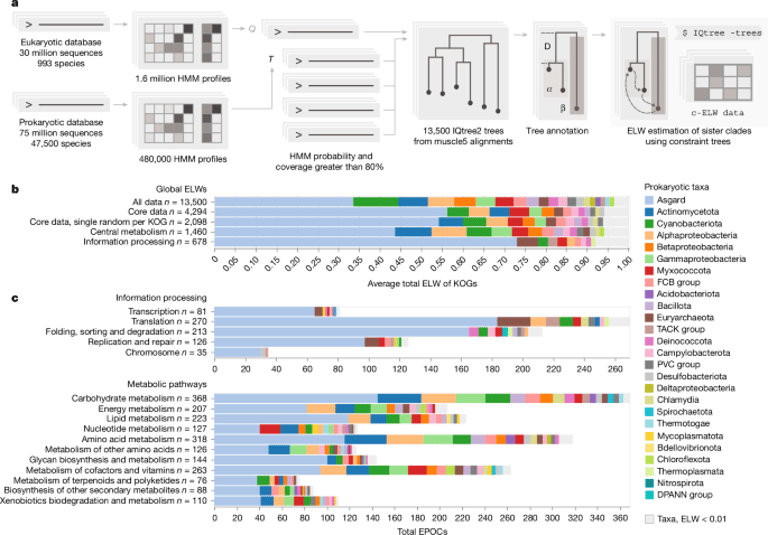
The study provides strong evidence that Asgard archaea were the dominant ancestral source for most core eukaryotic genes, supporting a view that Asgard ancestors largely shaped eukaryotic cellular complexity before LECA.
Findings support a model where many signature features of eukaryotic cells—developing within the Asgard lineage prior to LECA—were contributed by Asgard, with mitochondria arising from alphaproteobacterial endosymbiosis and subsequent integration of diverse bacterial genes.
Using a large, carefully filtered dataset of prokaryotic and eukaryotic sequences and a constrained-phylogeny approach, the authors find Asgard archaea most strongly associate with eukaryotic gene families across information-processing and many metabolic pathways, rather than other prokaryotes.
Beyond information processing, Asgard associations extend to nucleocytoplasmic transport, glycosylation, and protein targeting pathways, including ER-associated glycan biosynthesis and GPI-anchor synthesis, suggesting broad influence on membrane and protein-processing machinery.
Associations with Alphaproteobacteria are strongest in mitochondrial-related pathways, notably oxidative phosphorylation and iron-sulfur cluster biogenesis, implying a major contribution around the mitochondrial lineage with far less signal outside mitochondria.
Averaged ELW scores show strongest Asgard links to ribosome components, RNA/DNA polymerases, nucleotide excision repair, mismatch repair, homologous recombination, translation factors, protein targeting, and ubiquitin-proteasome–related processes, indicating Asgard ancestry across core information processing and substantial metabolism.
Lipid biosynthesis signals are strong for bacterial pathways affecting membrane biogenesis, with the highest Asgard signal in pathways tied to archaeal lipid precursors and eukaryotic mevalonate pathways, underscoring a nuanced, mosaic origin for lipid metabolism.
The study built 13,500 EPOCs from 10.9 million prokaryotic and 1.74 million eukaryotic sequences, using ELW to identify prokaryotic sister clades, yielding a core of about 4,290 high-quality EPOCs after filtering.
A substantial portion of eukaryotic genes show strongest associations with bacterial groups other than Asgard and Alphaproteobacteria, reflecting a mosaic, multi-prokaryotic ancestry for many LECA genes.
The dominant Asgard signal remains robust across different analytical representations, taxonomy frameworks, clustering strategies, and eukaryotic scope, and holds after quality control and dataset resampling, reinforcing the Asgard-dominated view of eukaryogenesis.
Summary based on 1 source
Get a daily email with more Science stories
Source

Nature • Jan 14, 2026
Dominant contribution of Asgard archaea to eukaryogenesis