CRISPRi Screen Maps Functional Enhancers in Astrocytes, Paving Way for Alzheimer's Precision Medicine
December 18, 2025
A large-scale CRISPRi enhancer screen in brain cells has produced a catalog of functional astrocyte enhancer regions, enabling interpretation of non-coding DNA variation in Alzheimer’s and other disorders.
The study lays groundwork for cell-type specific gene regulation and precision medicine approaches, while noting that clinical applications are not imminent.
The created dataset can train computational models and is already being used by DeepMind’s AlphaGenome project to benchmark enhancer-prediction models, potentially accelerating discovery.
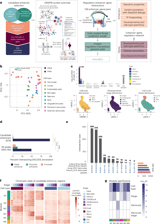
TT-seq data show that 73.1% of functional enhancers are transcribed, with bidirectional transcription and higher activity at functional enhancers.
Selected enhancers were validated with NanoString nCounter, qRT-PCR, and Western blot, confirming impact on both transcript and protein levels and concordance with CRISPRi results.
TF footprinting identifies 83 transcription factors enriched at functional enhancers, forming a core network of 18 astrocyte genes and 36 astrocyte TFs across 19 enhancers, supported by CRISPRi and TF–ChIP data.
Most enhancers regulate distal genes, with 76% acting within 200 kb and 50% within 50 kb; however, only about 38.6% target the nearest gene.
Enregulated genes are enriched for astrocyte-related processes, including chemotaxis, migration, wound response, and maturation/activation pathways, with 98 of 116 genes in at least one functional set.
Candidate enhancers were drawn from intergenic PsychENCODE regions, open astrocyte chromatin, and TADs around highly expressed genes; 61% were accessible in both fetal-derived and adult-derived astrocytes.
snATAC-seq across development shows astrocyte modules enriched for functional enhancers, with H3K27ac varying by cell type and module and H3K4me3 depleted at astrocyte enhancers.
Nearly 1,000 candidate enhancers were tested in human astrocytes using CRISPRi plus single-cell RNA sequencing to measure transcriptional impact.
The CRISPRi screen targeted 979 enhancers with 2,478 sgRNAs, identifying 158 enhancer–gene interactions that regulate 116 genes, with 84.1% causing downregulation and most enhancers affecting a single gene.
Summary based on 5 sources
Get a daily email with more Science stories
Sources

Medical Xpress • Dec 18, 2025
Hidden 'switches' in DNA reveal new insights into Alzheimer's disease
News-Medical • Dec 18, 2025
Junk' DNA may hold new clues to Alzheimer’s disease
Mirage News • Dec 18, 2025
Alzheimer's Clues Hidden in Junk DNA