Maternal Type 1 Diabetes May Epigenetically Shield Offspring from Autoimmunity, Study Finds
November 6, 2025
A follow-up project, supported with over $550,000 from The Leona M. and Harry B. Helmsley Charitable Trust, will identify which susceptibility genes are epigenetically modulated by maternal T1D, assess similar effects in gestational diabetes, and explore protein and metabolomic biomarkers across cohorts (GPPAD, BABYDIAB, BABYDIET, Fr1da).
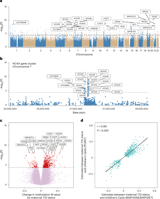
The study analyzed blood from 1,752 two-year-old children, comparing 790 with maternal T1D to 962 without, all at elevated genetic risk for T1D.
Methylation differences concentrate in regulatory regions (promoters, 5′ UTR, CpG islands) and show regulatory element enrichment across blood cells and tissues relevant to T1D (pancreas, thymus).
A Nature Metabolism study shows distinct DNA methylation changes in the blood of children exposed to maternal type 1 diabetes, pointing to epigenetic programming as a possible mechanism for protection against islet autoimmunity.
Differentially methylated CpGs correlate with gene expression (eQTM) for 142 genes, including several T1D susceptibility genes in the MHC region and HOXA genes, with 105 CpGs potentially regulating 15 T1D susceptibility genes.
The work builds on observations that offspring of mothers with T1D have lower T1D risk than offspring of affected fathers or siblings, offering mechanistic insight via epigenetic regulation.
A notable cluster of CpGs maps to HOXA genes on chromosome 7p15, especially HOXA5, indicating developmental gene regulation relevance; HOXA5 promoter regions show differential methylation validated by bisulfite pyrosequencing.
Compared with older offspring studies, there is limited overlap in exact CpG/DMR sites, but several shared target genes imply similar pathways.
Further work is needed to define the scope, significance, and practical implications of these epigenetic changes on offspring health and development.
Motif analysis reveals enrichment of MBD2 binding sites, a regulator of T cell development and regulatory T cell function, suggesting immune pathway involvement in the epigenetic changes.
The follow-up involves institutions across Europe, totaling funding of $550,000, to quantify protective epigenetic effects and extend analyses to gestational diabetes and multi-omics.
About 198 CpGs in offspring of mothers with T1D associate with 239 circulating protein traits, including CRISP2, a biomarker linked to progression to T1D.
The discussion emphasizes T1D's polygenic nature with MHC locus predominance, the potential roles of MBD2 and Treg regulation, and the HOXA/HOTAIRM1 axis in immune development, proposing targets for functional validation.
Findings align with risk variation by parental status, as risk is higher when fathers or siblings are affected, prompting exploration of maternal health effects during pregnancy.
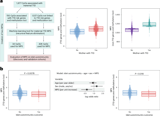
In offspring without maternal T1D, higher MPS at T1D-susceptibility loci correlates with reduced islet autoimmunity risk (OR 0.57 per unit MPS), a result replicated in the Fr1da cohort.
Type 1 diabetes involves autoimmune destruction of pancreatic beta cells requiring insulin, framing the significance of understanding epigenetic influences on disease risk.
Methylation propensity scores (MPS) were developed to capture maternal T1D–associated methylation patterns, with 34 CpGs linked to T1D susceptibility genes and 426 CpGs from non-susceptible loci, achieving AUCs of 0.71 and 0.88, respectively.
The study is presented as a preview of Nature Metabolism content, contextualizing environmental and familial factors influencing autoimmunity risk and linking to foundational epigenetic research.
Summary based on 6 sources
Get a daily email with more Science stories
Sources

News-Medical • Nov 6, 2025
Maternal type 1 diabetes confers a protective effect in children through epigenetic programming
Technology Networks • Nov 6, 2025
Maternal Diabetes Leaves Protective Marks on Children’s DNA