PLM-Interact: New AI Model Revolutionizes Protein Interaction Prediction, Outshines AlphaFold3
October 28, 2025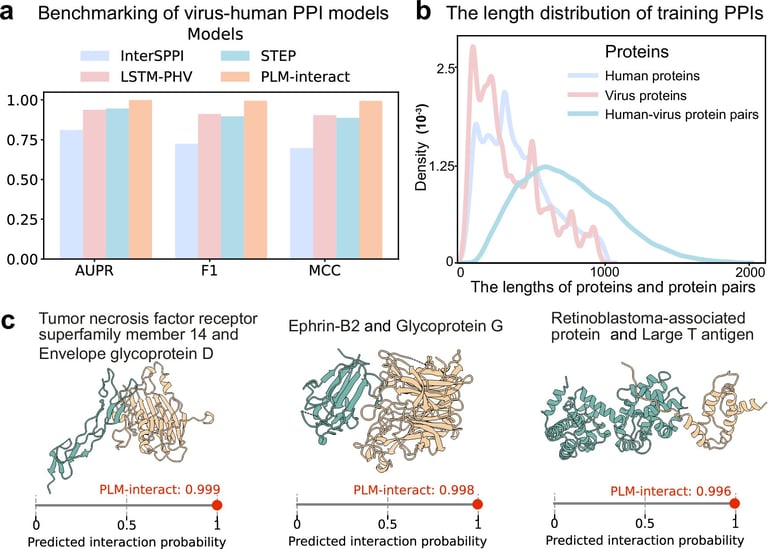
Researchers at the University of Glasgow have developed PLM-Interact, an advanced AI model trained on over 421,000 human protein pairs, which outperforms existing models by 16% to 28% in predicting protein interactions and can accurately identify key biological processes such as RNA polymerization and protein transport.
This model was built using the UK's DiRAC High Performance Supercomputer, capable of handling more than 650 million parameters, significantly speeding up the development process and enabling the handling of complex biological data.
The research underscores the importance of AI tools like PLM-Interact in responding to public health crises, such as COVID-19, by providing rapid insights into virus-host interactions and aiding in pandemic preparedness.
PLM-Interact represents a major step forward in creating highly accurate tools for predicting protein interactions, with promising applications in therapy and vaccine development.
The model can predict how mutations affect protein interactions, including those associated with genetic diseases and cancers, offering potential for therapeutic advancements and personalized medicine.
In tests involving over 22,000 human-virus protein interactions, PLM-Interact outperformed competitors like AlphaFold3, which only predicted one key interaction, demonstrating its potential in identifying emerging viruses and understanding viral mechanisms.
Supported by the EU Horizon 2020 program and UK research councils, the model shows promise in identifying disease-related mutations, aiding drug discovery, and predicting viral threats on a pandemic scale.
Further training expanded PLM-Interact’s scope to include an additional 22,383 interactions across human and viral proteins, enhancing its utility in understanding viral dynamics.
Developed by scientists at the University of Glasgow, PLM-Interact leverages astrophysical supercomputing resources to better understand protein-protein interactions and the effects of mutations, surpassing existing tools like AlphaFold3.
The model emphasizes the 'token layer'—the biological 'grammar'—to interpret protein behavior more accurately, reducing reliance on slow, costly experimental methods.
The research aims to accelerate biological discovery and plans to expand PLM-Interact’s capabilities to other disease areas such as virology and cancer biology, emphasizing its potential in medical research and pandemic response.
The development was supported by the DiRAC supercomputing facility, illustrating how infrastructure originally designed for astrophysics can be repurposed to advance biological research.
Summary based on 3 sources
Get a daily email with more AI stories
Sources

Phys.org • Oct 27, 2025
Supercomputer-developed AI learns the intricate language of biomolecules
SSBCrack News • Oct 27, 2025
University of Glasgow Scientists Develop Advanced AI Model to Decode Protein Interactions Using