Revolutionary Method ASCT Unveils True Antibiotic Effectiveness at Single-Cell Level, Transforming Treatment Approaches
January 9, 2026
ASCT across 405 Mycobacterium abscessus strains demonstrates that antibiotic killing is a genetically encoded trait.
Potential applications include personalizing antibiotic therapies to the infecting strain, accelerating tolerance tests, and enhancing evaluation of new drugs in clinical and industrial settings.
ASCT aligns more closely with real-world outcomes than animal studies or conventional clinical data, improving predictive power for treatment success.
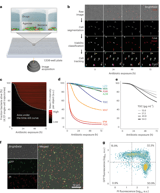
A new antimicrobial single-cell testing method determines whether antibiotics truly kill bacteria or merely inhibit growth, addressing a critical gap in traditional assays. This method, ASCT, measures bacterial killing at single-cell resolution and scales to millions of cells in 1,536-well plates, linking in vitro killing dynamics to real infection outcomes.
Genetic analyses show drug tolerance in M. abscessus is heritable and can be clonal or parallel-evolved, with certain lineages exhibiting clinically relevant tolerance patterns, such as tigecycline tolerance, arising from genetic variation.
Starvation-based killing assays achieve high predictive accuracy for regimen efficacy in TB models, suggesting starvation-tolerant populations play a key role in long-term outcomes.
Led by Dr. Lucas Boeck at the University of Basel and University Hospital Basel, the findings were published in Nature Microbiology in 2026.
Researchers tracked over 140 million mycobacteria and analyzed roughly 20,000 time–kill curves to identify determinants of antibiotic killing and its clinical relevance.
The method was validated with 65 drug combinations against Mycobacterium tuberculosis and analyzed samples from about 400 patients with Mycobacterium abscessus, revealing both therapy-to-therapy and strain-to-strain differences in kill effectiveness.
ASCT reveals substantial strain-to-strain variability in time–kill kinetics under eight antibiotics in Mycobacterium abscessus, showing that genetic background modulates killing and survival even with identical drug exposure.
In tuberculosis experiments, killing under starvation conditions better predicted regimen efficacy in mice and humans than traditional CFU/MIC measures, indicating non-replicating bacteria drive outcomes.
ASCT uses propidium iodide to mark dead cells and time-lapse imaging to build time–kill curves that capture population-level killing, tolerance, and persistence across regimens.
Summary based on 4 sources
Get a daily email with more Science stories
Sources
Phys.org • Jan 9, 2026
Single-cell testing shows which antibiotics actually kill bacteria, not just stop growth
GEN - Genetic Engineering and Biotechnology News • Jan 9, 2026
New Single‑Cell Testing Measures How Effectively Antibiotics Kill Bacteria
SciTechDaily • Jan 9, 2026
Some Antibiotics Don’t Kill Bacteria. This Test Shows Which Do